A Deep Learning Pipeline for Assessing Ventricular Volumes from a Cardiac MRI Registry of Patients with Single Ventricle Physiology
Deep Learning Pipeline for Assessing Ventricular Volumes from Cardiac MRI in Single Ventricle Physiology
Introduction/Abstract
The aim of the paper is to develop an end-to-end deep learning pipeline for:
- Automated ventricular segmentation of cardiac MRI scans
- Providing cardiac function metrics for a cardiac MRI registry of patients with single ventricular physiology
Evaluation metric: Dice Score
Result analysis: Bland-Altman and intraclass correlation analysis
Results:
- Dice: 0.91 for end diastolic
- Dice: 0.86 for end-systolic
- Dice: 0.74 for myocardium
The pipeline was further tested on 475 unseen patient examinations.
Dataset Overview: Fontan Outcomes Registry Using CMR Examinations (FORCE): >4500 scans from >3000 patients from a multicentre cardiac MRI registry of patients who have undergone the Fontan palliation.
The issue with the dataset is that it is not standardised and it is not feasible to perform standardised manual segmentation of all scans again.
The pipeline automatically identifies ventricular short-axis cine stacks, crops out the heart, and segments the ventricles of patients with Fontan circulation.
Study objectives:
- Develop and validate each step in the pipeline on a curated dataset with ground truth segmentations
- Test the pipeline on large amounts of registry data, both in terms of segmentation quality and comparison with the volumes from the original reports
Dataset Details
250 samples from the FORCE registry consisting of samples from 13 institutes:
- Training: 175
- Validation: 25
- Testing: 50
Data was collected between November 2007–December 2022, consisting of scans from both 1.5T and 3T MRI scanners.
Conditions for inclusion include:
- Existence of short-axis cine stacks
- At least 6 slices per stack
- At least 10 frames per cardiac cycle to be categorised as a cine
- All relevant DICOM header information is present
Ground Truth Label Generation:
- Single clinical researcher performed initial segmentations
- Three cardiovascular imaging physicians reviewed and corrected 150 cases
- Standardisation protocol that was used: trabeculae and papillary muscles included in blood pool, underdeveloped secondary ventricles contoured (important as this differs from institution to institution in the FORCE registry)
Methodology
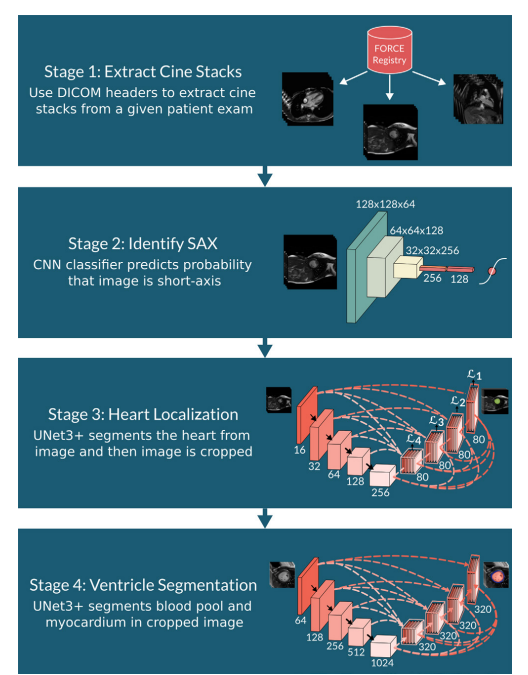
There are 4 stages in the pipeline:
- Stage 1: Extract the cine stacks
- Stage 2: Identify whether the image is Short Axis (SAX)
- Stage 3: Localise the heart and segment using a UNet3+
- Stage 4: Ventricle Segmentation
Stage 1: Cine Stack Extraction
Each entry in the registry is an examination and thus contains a lot of information and thousands of images. The paper shows examples ranging from 460 to 21,000 files per patient, organised into 5–290 different series. These files include:
- Different anatomical views (short-axis, long-axis, four-chamber)
- Static images vs. moving sequences (cines)
- Different orientations and slice positions
- Various pulse sequences and contrasts
DICOM headers are used to group the images that belong together by checking information such as:
- Orientation
- Pixel size
- Slice thickness
- At least 10 frames (that makes a cine)
- At least 6 slices (that makes it a stack)
Stage 2: Short Axis Identification
The short-axis views consist of cross-sectional views of the heart. The total volume can be measured by stacking the slices from base to apex and then adding up the slice areas. SAX offers complete coverage of the ventricles and provides geometric simplicity (cross-sections are circular).
Stage 3: Heart Localisation and Cropping
Use a UNet3+ model to localise the blood pool and subsequently crop the heart ou
Stage 4: Ventricular Segmentation
Results
SAX Identification
SAX identification per slice:
- Accuracy: 96.1%
- Precision: 98%
- Recall: 94.4%
Since all slices are considered when making the final classification, all 50 test samples were correctly identified.
Heart Localisation
Median IoU: 0.94
Ventricle Segmentation Model
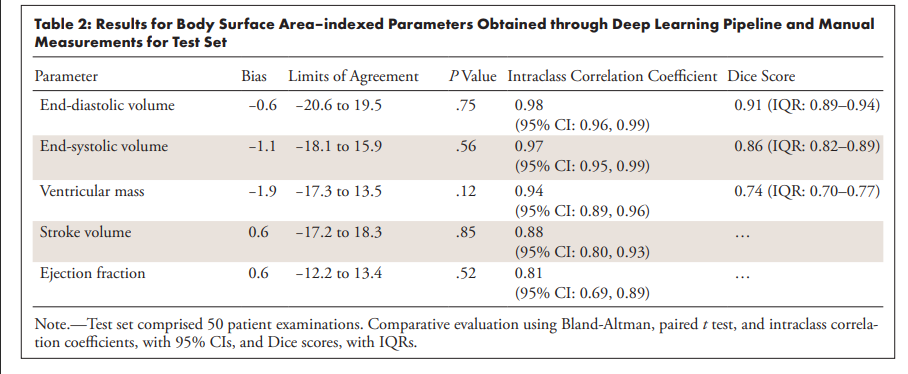
Dice scores:
- Blood Pool (Diastole): 0.91
- Blood Pool (Systole): 0.86
- Ventricular Mass: 0.74
Volume Measurements (Difference between AI and Human Measurements):
- End Diastolic Volume (EDV): 0.6 mL/m²
- End Systolic Volume (ESV): 1.1 mL/m²
- Stroke Volume: 0.6 mL/m²
- Ejection Fraction: 0.6%
- Ventricular Mass: 1.9 g/m²
Analysis
Category: A clinical application research applying deep learning to automate ventricular volume assessment from cardiac MRI in patients with single ventricle physiology (Fontan circulation).
Context: Addresses a critical gap in congenital heart disease care where patients with functionally single ventricles require lifelong cardiac monitoring via MRI. The FORCE registry contains >4500 cardiac MRI scans from >3000 patients, but the volumetric data from original clinical reports are unreliable due to different segmentation protocols and interobserver variability. Manual core laboratory segmentation of the entire registry is unfeasible, hence automated methods should be considered and developed.
Correctness: The methodology appears robust with a well-designed four-stage pipeline. The validation approach is thorough, using 250 patients for training/validation/testing with ground truth segmentations reviewed by experienced physicians. However, manual segmentations were performed by a single operator.
Contributions:
- First end-to-end deep learning pipeline for automated segmentation of single ventricular physiology
- Handles extremely heterogeneous anatomy from 13+ institutions with varying protocols
- Achieves high accuracy comparable to manual segmentation (Dice scores >0.86 for blood pool)
- Demonstrates feasibility for large-scale registry processing (475 unseen cases in <40 hours)
Clarity: Well-structured with clear methodology. The four-stage pipeline is logically presented, though the extensive technical details and statistical analyses make some sections dense.
Main Task
The paper develops an automated deep learning pipeline to segment cardiac ventricles from MRI scans in patients with single ventricle physiology (those who've had Fontan surgery). The pipeline automatically:
- Extracts relevant image sequences from MRI studies
- Identifies short-axis cardiac views
- Crops the heart region
- Segments ventricular blood pool and muscle tissue
- Calculates clinical metrics (volumes, ejection fraction, etc.)
Results Explanation
Technical Performance:
- Dice scores measure overlap between automated and manual segmentations (1.0 = perfect match):
- End-diastolic blood pool: 0.91 (excellent)
- End-systolic blood pool: 0.86 (very good)
- Myocardium: 0.74 (good)
Clinical Metrics Agreement: The automated measurements showed excellent agreement with manual measurements:
- End-diastolic volume bias: -0.6 mL/m² (minimal difference)
- End-systolic volume bias: -1.1 mL/m² (minimal difference)
- Intraclass correlation coefficients: >0.97 for volumes (near-perfect reliability)
Large-Scale Testing: When applied to 475 unseen cases:
- 68% required no adjustments (satisfactory)
- 26% needed minor adjustments
- 5% needed major adjustments
- 0.4% failed completely
Clinical Relevance: The automated measurements disagreed significantly with original clinical reports, but so did manual core laboratory measurements. This suggests the original clinical data were highly variable due to different institutional protocols, highlighting the need for standardised automated processing.
Practical Impact: The pipeline processes each patient in ~26 seconds, making it feasible to reprocess the entire 4500+ examination registry in under 40 hours, providing standardised, reliable cardiac function data for research and clinical decision-making.